Modelos de Clasificación
1º Congreso Latinoamericano de Mujeres en Bioinformática y Ciencia de Datos
1 / 43
Vamos a modelar las especies 🐧
Ingreso los datos
library(tidymodels) library(palmerpenguins)penguins <- palmerpenguins::penguins %>% drop_na() %>% #elimino valores perdidos select(-year,-sex, -island) #elimino columnas q no son numéricasglimpse(penguins) #observo variables restantes## Rows: 333## Columns: 5## $ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Ade…## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, 36.7, 39.3, 38.9, 39.2, 41.1, 38.…## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, 19.3, 20.6, 17.8, 19.6, 17.6, 21.…## $ flipper_length_mm <int> 181, 186, 195, 193, 190, 181, 195, 182, 191, 198, 1…## $ body_mass_g <int> 3750, 3800, 3250, 3450, 3650, 3625, 4675, 3200, 380…2 / 43
Paso 1: Vamos a dividir el set de datos
library(rsample)set.seed(123) #setear la semillap_split <- penguins %>% initial_split(prop=0.75) # divido en 75%p_train <- training(p_split)p_split## <Analysis/Assess/Total>## <250/83/333># para hacer validación cruzada estratificadap_folds <- vfold_cv(p_train, strata = species)Estos son los datos de entrenamiento/prueba/total
- Vamos a entrenar con 250 muestras
- Vamos a testear con 83 muestras
- Datos totales: 333
3 / 43
Paso 2: Preprocesamiento (receta)
#creo la recetarecipe_dt <- p_train %>% recipe(species~.) %>% step_corr(all_predictors()) %>% #elimino las correlaciones step_center(all_predictors(), -all_outcomes()) %>% #centrado step_scale(all_predictors(), -all_outcomes()) %>% #escalado prep() recipe_dt #ver la receta## Data Recipe## ## Inputs:## ## role #variables## outcome 1## predictor 4## ## Training data contained 250 data points and no missing data.## ## Operations:## ## Correlation filter removed no terms [trained]## Centering for bill_length_mm, ... [trained]## Scaling for bill_length_mm, ... [trained]4 / 43
Paso 3: Especificar el modelo 🌳
Modelo de árboles de decisión
Vamos a utilizar el modelo por defecto
#especifico el modelo set.seed(123)vanilla_tree_spec <- decision_tree() %>% #arboles de decisión set_engine("rpart") %>% #librería rpart set_mode("classification") #modo para clasificarvanilla_tree_spec## Decision Tree Model Specification (classification)## ## Computational engine: rpart5 / 43
Paso 4: armo el workflow
#armo el workflowtree_wf <- workflow() %>% add_recipe(recipe_dt) %>% #agrego la receta add_model(vanilla_tree_spec) #agrego el modelotree_wf## ══ Workflow ═══════════════════════════════════════════════════════════════════════════════════════════## Preprocessor: Recipe## Model: decision_tree()## ## ── Preprocessor ───────────────────────────────────────────────────────────────────────────────────────## 3 Recipe Steps## ## ● step_corr()## ● step_center()## ● step_scale()## ## ── Model ──────────────────────────────────────────────────────────────────────────────────────────────## Decision Tree Model Specification (classification)## ## Computational engine: rpart6 / 43
Paso 5: Ajuste de la función
#modelo vanilla sin tunningset.seed(123) vanilla_tree_spec %>% fit_resamples(species ~ ., resamples = p_folds) %>% collect_metrics() #desanidar las metricas## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 accuracy multiclass 0.953 10 0.0154## 2 roc_auc hand_till 0.946 10 0.01977 / 43
Paso 5.1: vamos a especificar 2 hiperparámatros
set.seed(123) trees_spec <- decision_tree() %>% set_engine("rpart") %>% set_mode("classification") %>% set_args(min_n = 20, cost_complexity = 0.1) #especifico hiperparámetrostrees_spec %>% fit_resamples(species ~ ., resamples = p_folds) %>% collect_metrics()## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 accuracy multiclass 0.953 10 0.0154## 2 roc_auc hand_till 0.946 10 0.01978 / 43
Ejercicio
1. ¿Por qué es el mismo valor obtenido en los dos casos?
2. Dejando fijo el valor de min_n=20, pruebe C=1, C=0.5 y C=0.
3. Dejando fijo el valor de C=0, pruebe min_n 1 y 5.
05:00
9 / 43
Paso 6: Predicción del modelo 🔮
#utilizamos la funcion last_fit junto al workflow y al split de datosfinal_fit_dt <- last_fit(tree_wf, split = p_split)final_fit_dt %>% collect_metrics()## # A tibble: 2 x 3## .metric .estimator .estimate## <chr> <chr> <dbl>## 1 accuracy multiclass 0.916## 2 roc_auc hand_till 0.92910 / 43
Paso 6.1: matriz de confusión
final_fit_dt %>% collect_predictions() %>% conf_mat(species, .pred_class) #para ver la matriz de confusión## Truth## Prediction Adelie Chinstrap Gentoo## Adelie 33 2 0## Chinstrap 2 16 0## Gentoo 2 1 2711 / 43
Paso 6.1: matriz de confusión
final_fit_dt %>% collect_predictions() %>% conf_mat(species, .pred_class) #para ver la matriz de confusión## Truth## Prediction Adelie Chinstrap Gentoo## Adelie 33 2 0## Chinstrap 2 16 0## Gentoo 2 1 27final_fit_dt %>% collect_predictions() %>% sens(species, .pred_class) #sensibilidad global del modelo## # A tibble: 1 x 3## .metric .estimator .estimate## <chr> <chr> <dbl>## 1 sens macro 0.91111 / 43
Ejercicio
Repetir estos pasos para el modelo de C=0 y min_n=5
05:00
12 / 43
Resumiendo
Paso 1:
Dividimos los datos
- initial_split()

13 / 43
Resumiendo
Paso 2:
Preprocesamiento de los datos
- step_*()

14 / 43
Resumiendo
Paso 3:
Especificamos el modelo y sus args
- set_engine()
- mode()

15 / 43
Resumiendo
Paso 4:
Armamos el workflow con la receta y el modelo
- workflow()
- add_recipe()
- add_model()

16 / 43
Resumiendo
Paso 5: Tune
Tuneo de los hiperparámetros
- last_fit()

17 / 43
Resumiendo
Paso 6:
Predicción y comparación de las métricas
- collect_metrics()
- collect_predictions() + conf_mat()

18 / 43
Random Forest
19 / 43
Paso 2: Preprocesamos los datos
p_recipe <- training(p_split) %>% recipe(species~.) %>% step_corr(all_predictors()) %>% step_center(all_predictors(), -all_outcomes()) %>% step_scale(all_predictors(), -all_outcomes()) %>% prep()p_recipe## Data Recipe## ## Inputs:## ## role #variables## outcome 1## predictor 4## ## Training data contained 250 data points and no missing data.## ## Operations:## ## Correlation filter removed no terms [trained]## Centering for bill_length_mm, ... [trained]## Scaling for bill_length_mm, ... [trained]20 / 43
Paso 3: Especificar el modelo
rf_spec <- rand_forest() %>% set_engine("ranger") %>% set_mode("classification")21 / 43
Veamos como funciona el modelo sin tunning
set.seed(123)rf_spec %>% fit_resamples(species ~ ., resamples = p_folds) %>% collect_metrics()## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 accuracy multiclass 0.972 10 0.0108 ## 2 roc_auc hand_till 0.996 10 0.0019222 / 43
Random Forest con mtry=2 🔧
rf2_spec <- rf_spec %>% set_args(mtry = 2)set.seed(123)rf2_spec %>% fit_resamples(species ~ ., resamples = p_folds) %>% collect_metrics()## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 accuracy multiclass 0.972 10 0.0108 ## 2 roc_auc hand_till 0.996 10 0.0019223 / 43
Random Forest con mtry=3 🔧
rf3_spec <- rf_spec %>% set_args(mtry = 3)set.seed(123)rf3_spec %>% fit_resamples(species ~ ., resamples = p_folds) %>% collect_metrics()## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 accuracy multiclass 0.967 10 0.0104 ## 2 roc_auc hand_till 0.997 10 0.0011524 / 43
Random Forest con mtry=4 🔧
rf4_spec <- rf_spec %>% set_args(mtry = 4)set.seed(123)rf4_spec %>% fit_resamples(species ~ ., resamples = p_folds) %>% collect_metrics()## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 accuracy multiclass 0.964 10 0.00972## 2 roc_auc hand_till 0.996 10 0.0014725 / 43
Tuneo de hiperparámetros automático 🔧
tune_spec <- rand_forest( mtry = tune(), trees = 1000, min_n = tune()) %>% set_mode("classification") %>% set_engine("ranger")tune_spec## Random Forest Model Specification (classification)## ## Main Arguments:## mtry = tune()## trees = 1000## min_n = tune()## ## Computational engine: ranger26 / 43
Workflows
tune_wf <- workflow() %>% add_recipe(p_recipe) %>% add_model(tune_spec)set.seed(123)cv_folds <- vfold_cv(p_train, strata = species)tune_wf## ══ Workflow ═══════════════════════════════════════════════════════════════════════════════════════════## Preprocessor: Recipe## Model: rand_forest()## ## ── Preprocessor ───────────────────────────────────────────────────────────────────────────────────────## 3 Recipe Steps## ## ● step_corr()## ● step_center()## ● step_scale()## ## ── Model ──────────────────────────────────────────────────────────────────────────────────────────────## Random Forest Model Specification (classification)## ## Main Arguments:## mtry = tune()## trees = 1000## min_n = tune()## ## Computational engine: ranger27 / 43
Paralelizamos los cálculos
doParallel::registerDoParallel()set.seed(123)tune_res <- tune_grid( tune_wf, resamples = cv_folds, grid = 20)tune_res## # Tuning results## # 10-fold cross-validation using stratification ## # A tibble: 10 x 4## splits id .metrics .notes ## <list> <chr> <list> <list> ## 1 <split [224/26]> Fold01 <tibble [40 × 6]> <tibble [0 × 1]>## 2 <split [224/26]> Fold02 <tibble [40 × 6]> <tibble [0 × 1]>## 3 <split [224/26]> Fold03 <tibble [40 × 6]> <tibble [0 × 1]>## 4 <split [224/26]> Fold04 <tibble [40 × 6]> <tibble [0 × 1]>## 5 <split [224/26]> Fold05 <tibble [40 × 6]> <tibble [0 × 1]>## 6 <split [225/25]> Fold06 <tibble [40 × 6]> <tibble [0 × 1]>## 7 <split [225/25]> Fold07 <tibble [40 × 6]> <tibble [0 × 1]>## 8 <split [226/24]> Fold08 <tibble [40 × 6]> <tibble [0 × 1]>## 9 <split [227/23]> Fold09 <tibble [40 × 6]> <tibble [0 × 1]>## 10 <split [227/23]> Fold10 <tibble [40 × 6]> <tibble [0 × 1]>28 / 43
Rango de valores para min_n y mtry
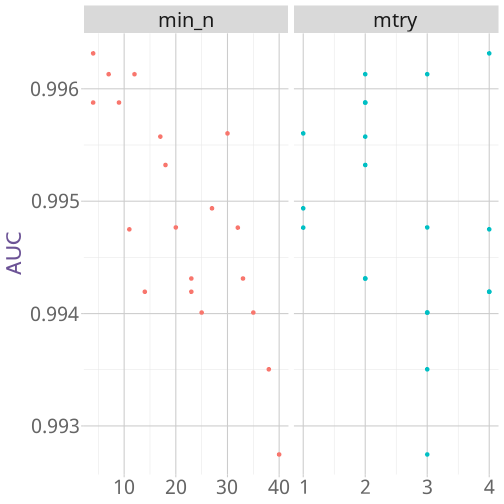
29 / 43
Elijo el mejor modelo 🏆
- Con la función select_best
best_auc <- select_best(tune_res, "roc_auc")final_rf <- finalize_model( tune_spec, best_auc)final_rf## Random Forest Model Specification (classification)## ## Main Arguments:## mtry = 4## trees = 1000## min_n = 4## ## Computational engine: ranger30 / 43
Valores finales
set.seed(123)final_wf <- workflow() %>% add_recipe(p_recipe) %>% add_model(final_rf)final_res <- final_wf %>% last_fit(p_split)final_res %>% collect_metrics()## # A tibble: 2 x 3## .metric .estimator .estimate## <chr> <chr> <dbl>## 1 accuracy multiclass 0.952## 2 roc_auc hand_till 0.99931 / 43
Matriz de Confusión
final_res %>% collect_predictions() %>% conf_mat(species, .pred_class)## Truth## Prediction Adelie Chinstrap Gentoo## Adelie 39 0 1## Chinstrap 2 11 1## Gentoo 0 0 2932 / 43
Ejercicio 3
Con el set de datos de iris realice una clasificación con random forest.
10:00
33 / 43
Resumiendo
Paso 2: Recipes
Preprocesamiento de los datos

34 / 43
Resumiendo
Paso 3: Parsnip
Especificamos el modelo y sus args

35 / 43
Resumiendo
Paso 4: Workflow
Armamos el workflow con la receta y el modelo

36 / 43
Resumiendo
Paso 5: Tune
Tuneo de los hiperparámetros

37 / 43
Resumiendo
Paso 6:
Predicción y comparación de las métricas

38 / 43
Importancia de las variables
- libreria vip
library(vip)set.seed(123)final_rf %>% set_engine("ranger", importance = "permutation") %>% fit(species ~ ., data = juice(p_recipe)) %>% vip(geom = "point")+ theme_xaringan()39 / 43
Gráfico
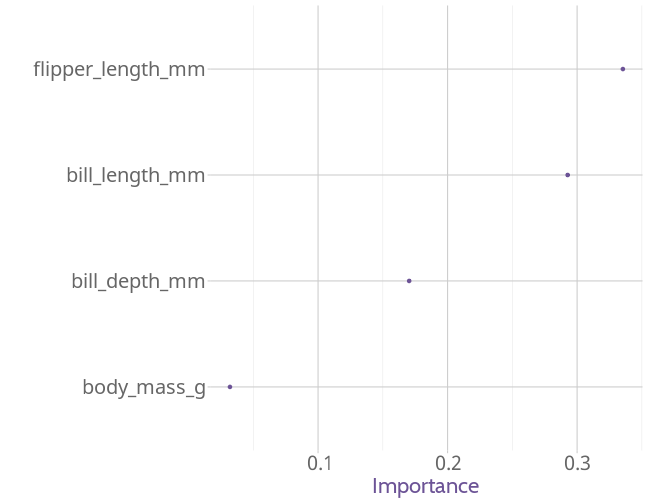
40 / 43
Ejemplos (un poco) más reales
Tuning random forest hyperparameters with #TidyTuesday trees data
Hyperparameter tuning and #TidyTuesday food consumption
41 / 43
Bibliografía
Max Kuhn & Julia Silge - Tidy Modeling (en desarrollo)
Julia Silge's Personal Blog
Max Kuhn & Kjell Johnson - Feature engineering and Selection: A Practical Approach for Predictive Models
Max Kuhn & Kjell Johnson - Applied Predictive Modeling
Documentación de tidymodels
Alison Hill rstudio conf y curso virtual material
42 / 43
Muchas gracias 🤖
43 / 43